Numerical 1-D FEM solution of the heat diffusion equation
[90]:
import numpy as np
import matplotlib.pyplot as plt
import sympy as sy
from scipy.sparse import csr_matrix
from scipy import sparse
from numpy import array
from scipy.sparse.linalg import spsolve
import matplotlib as mpl
mpl.rcParams['figure.dpi']= 300
from tabulate import tabulate
First we define some numerical constants and define the problem
[91]:
#constants
lx = 10 #width
nnod = 11 #number of nodes
nnodel = 2 #nodes per element
k = 1 #conductivity
Q = 1 #source term
Tleft = 0 #T boundary conditions
Trite = 0
#numerics
nel = nnod-1 #numnber of elements
Now we make our FEM mesh and element connectivity
[92]:
Gcoord, dx = np.linspace(0,lx,nnod, retstep=True) #global coordinates
EL2NOD = np.array([np.arange(0,nnod-1), np.arange(1,nnod)], dtype=int).T.copy() # connectivity matrix
print(tabulate(EL2NOD, headers=['Element #', 'Node 1', 'Node 2'], showindex=True))
Element # Node 1 Node 2
----------- -------- --------
0 0 1
1 1 2
2 2 3
3 3 4
4 4 5
5 5 6
6 6 7
7 7 8
8 8 9
9 9 10
Now we spell out our analytically-derived element stiffness matrix (see previous sections):
[84]:
#Ael = ???
#Rhs_el = ???
[104]:
# hide
Ael = np.array([[k/dx, -k/dx],[-k/dx, k/dx]])
Rhs_el = np.array([Q*dx/2, Q*dx/2])
Now we need to loop over all elements and assemble the global stiffness matrix
[100]:
Rhs_all = np.zeros(nnod)
I = np.zeros((nel,nnodel*nnodel))
J = np.zeros((nel,nnodel*nnodel))
K = np.zeros((nel,nnodel*nnodel))
for iel in range(0,nel):
I[iel,:] = (EL2NOD[iel,:]*np.ones((nnodel,1), dtype=int)).T.reshape(nnodel*nnodel)
J[iel,:] = (EL2NOD[iel,:]*np.ones((nnodel,1), dtype=int)).reshape(nnodel*nnodel)
K[iel,:] = Ael.reshape(nnodel*nnodel)
Rhs_all[EL2NOD[iel,:]] += Rhs_el
A_all = sparse.csr_matrix((K.reshape(nel*nnodel*nnodel),(I.reshape(nel*nnodel*nnodel),J.reshape(nel*nnodel*nnodel))),shape=(nnod,nnod))
Now we apply the boundary conditions in the usual way and solve using a sparse solver!
[101]:
#Apply boundary conditions
#A_all??
#A_all??
#A_all??
# A_all??
#Rhs_all[0] = ?
#Rhs_all[-1] = ?
[102]:
# hide
#Apply boundary conditions
A_all[0,0] = 1
A_all[0,1] = 0
A_all[nnod-1,nnod-1] = 1
A_all[nnod-1,nnod-2] = 0
Rhs_all[0] = Tleft
Rhs_all[-1] = Trite
[103]:
# and solve!
Tnew=spsolve(A_all,Rhs_all)
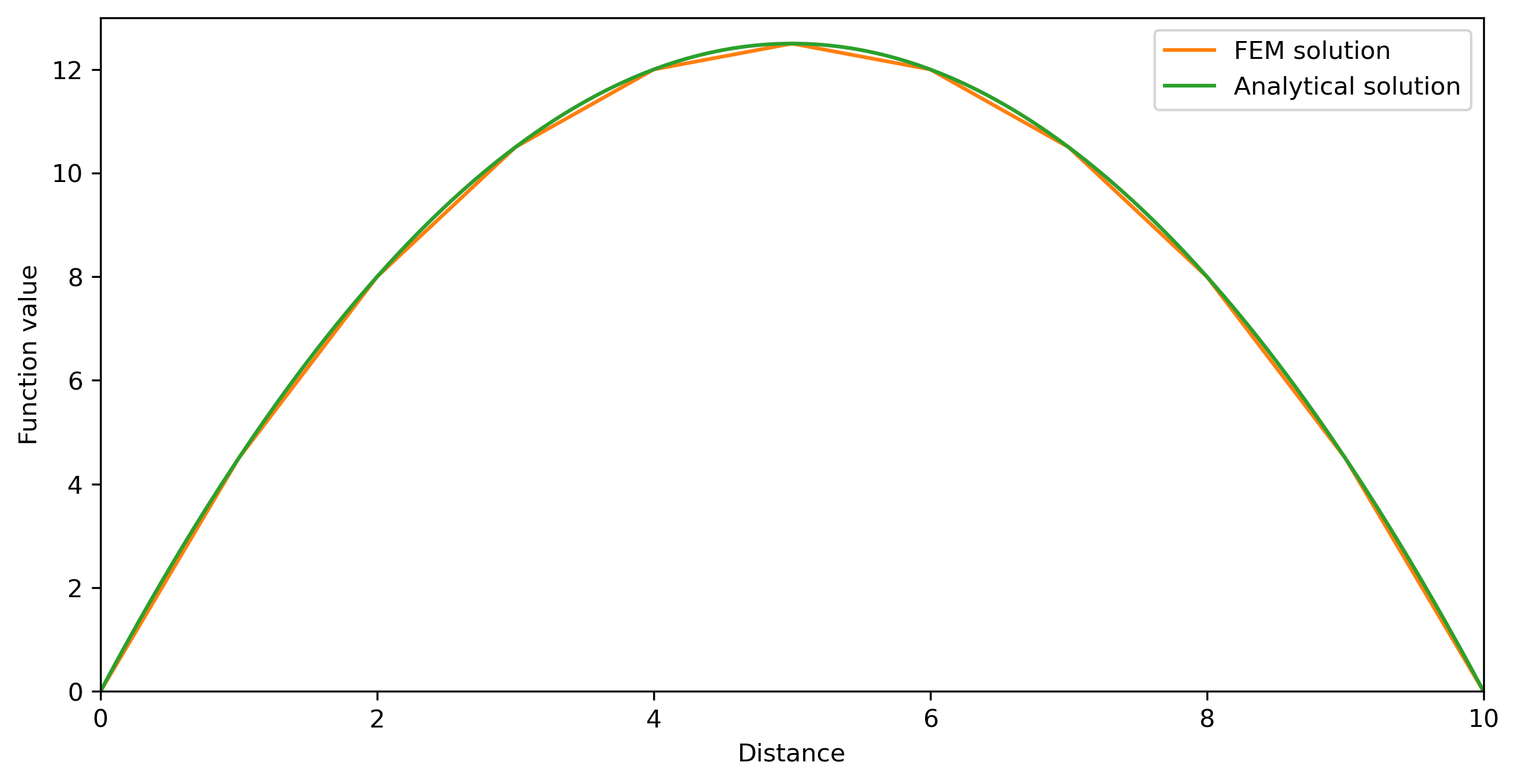
#Analytical solution
X = np.linspace(0,lx, 100*nnod)
T_ana = -1/2*Q/k*np.square(X) + 1/2*Q/k*lx*X;
# Plotting
fig = plt.figure(figsize=(10,5))
fig.clf()
ax = plt.axes(xlim=(0, lx), ylim=(0, 13))
line, = ax.plot([], [], lw=1)
ax.set_xlabel('Distance')
ax.set_ylabel('Function value')
plt.plot(Gcoord, Tnew, label='FEM solution')
plt.plot(X, T_ana, label='Analytical solution')
plt.legend()
[103]:
<matplotlib.legend.Legend at 0x7f86c16d0890>
[ ]: